EyeNet Code Metrics:
Accuracy (Training): 82%
Accuracy (Testing): 80%
Precision: 88%


EyeNet Code Metrics:
Accuracy (Training): 82%
Accuracy (Testing): 80%
Precision: 88%
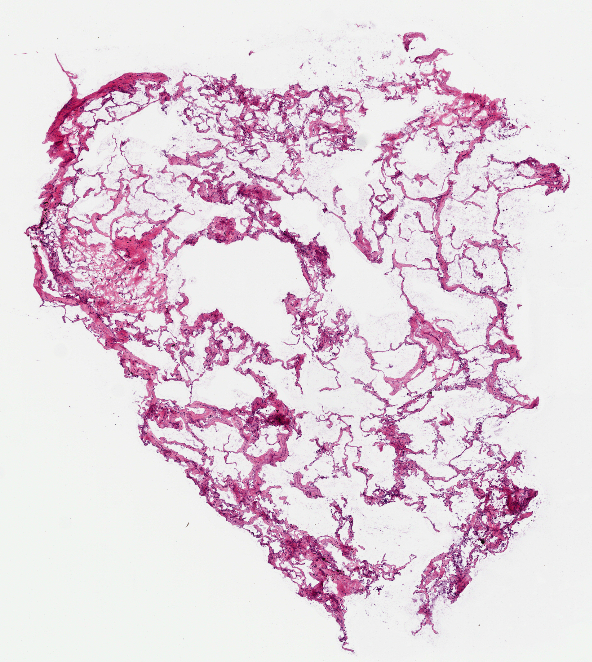


While EyeNet utilizied downsampled images of eyes into 256 by 256 squares, the data I attempted to use would be medical scan files (in the .svs format) which were 10s of Mb even in compressed form, which took roughly a minute to download per slide. While evidence of Diabetic Retinopathy in eye pictures was preserved for the most part even after this downsampling, I was worried that the details of cancer scan files would need to be preserved in close to their entirety. Combined with the inability to download identical tissue scans and cancer types in large numbers, it seemed almost all training data would have to be hand-selected.
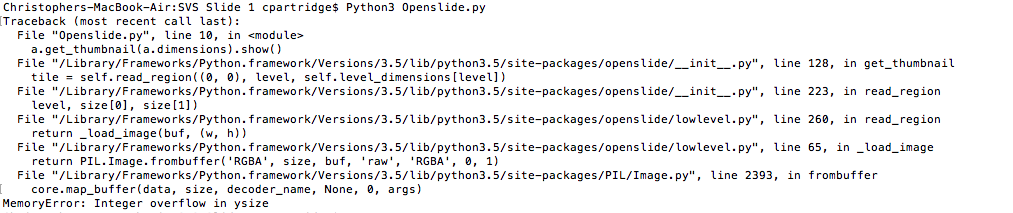
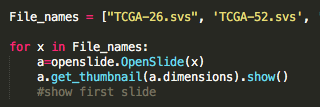
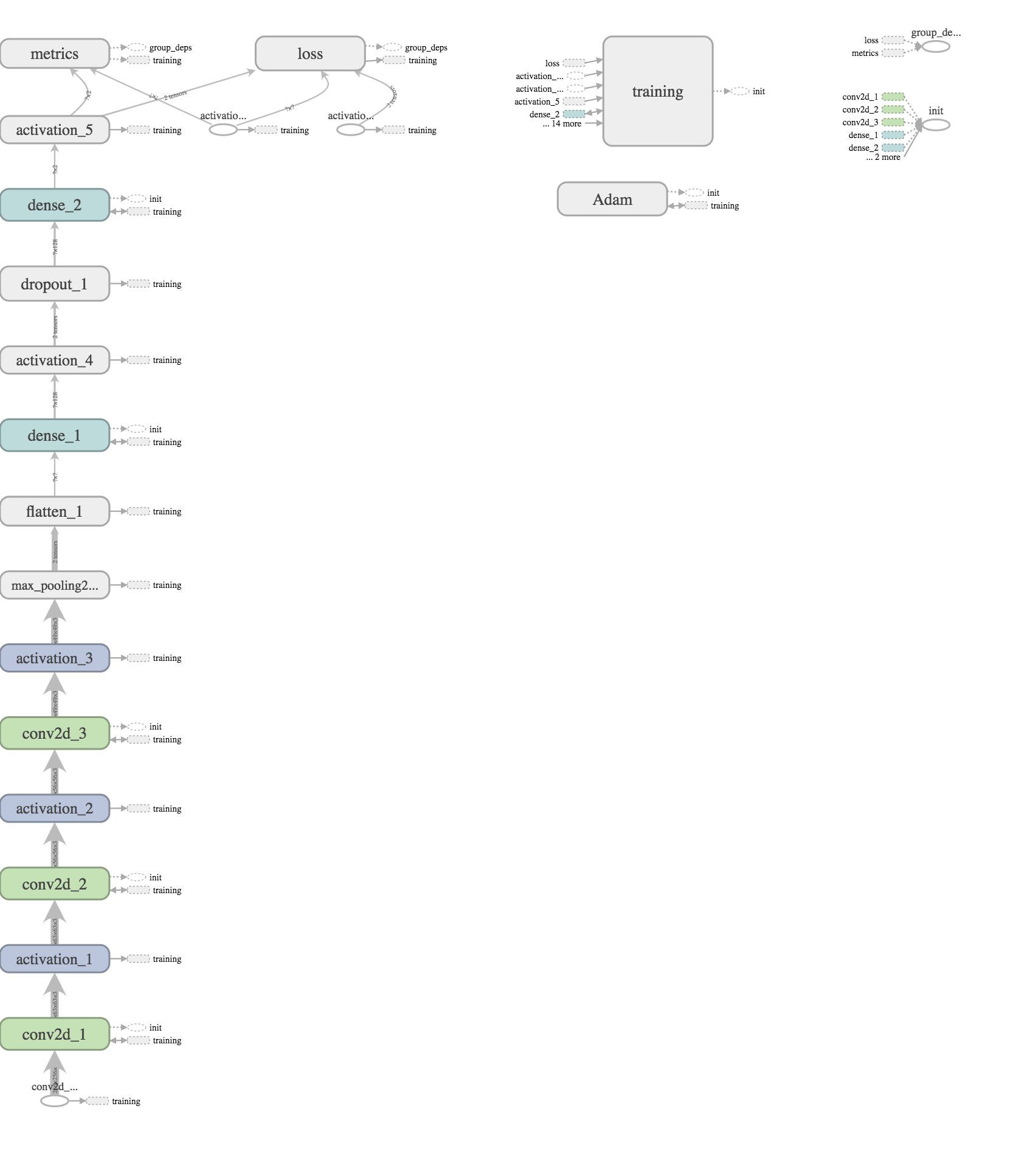
Because the file format was not readily accessible, I had to implement Python API of OpenSlide link) , and write code to extract the files as well as their annotations to begin training the correlation of the two for image classification. The goal from there would be to take scans of non-cancerous cells as well as cancerous cells of the same tissue and the same cancer, leaving the dense, 128 layer CNN to correlate simply cancerous cells vs. not. However, even in extracting the files for programatic manipulation, I ran into overflow issues.
Image (left to right): Breast Cancer cell, Lung Carcinoma cell (1), Lung Carcinoma (2) Extraction code lines, Error in image extraction.
